New Members
New members like Prof. Sánchez-García or Prof. Probst introduce new research interests to the CCSS; especially in the initial stages, they need the Center’s support with administration, computer use and, in particular, computing time resources.
Dr. Alexander Probst is head of the Group for Aquatic Microbial Ecology (GAME) at the Biofilm Center of the University of Duisburg-Essen, where he is also professor of Aquatic Microbial Ecology. Prof. Probst transferred to Essen from the University of California, Berkeley, where he conducted research on cold-water geysers as a postdoc. He is now continuing this research at the UDE, with a special focus on the deep terrestrial subsurface biosphere, which is largely populated by unknown microorganisms. These unfamiliar life forms are being classified in the course of the research in the “tree of life” (Hug et al. 2016) – a constantly evolving research and teaching model. Genome sequencing means that new organisms in the environment are being discovered almost daily, all of which require taxonomic classification. For some groups this can be a quick process, provided that relevant neighbouring groups are already known. When it comes to entirely new species for which there are no known closely related organisms, however, classification can be more of a challenge. The research group works on metabolic processes in the terrestrial subsurface, more precisely the deep biosphere. This is an environment in which almost every sample reveals countless new microbial species for classification. A new article published in Nature Microbiology, for example, presents over 1,200 new genomes. To continue expanding the tree of life and correctly classify new organisms, Alexander Probst and his group at the UDE need around 300,000 hours of CPU time a year. The algorithms they use include maximum likelihood methods and Bayesian inferences to calculate phylogenetic trees based on alignments of concatenated marker genes. The results will be used in very many specialist articles that will come out of the research group.
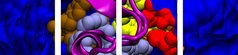
Prof. Sánchez-García’s field of research is computational chemistry and computational chemical biology, with emphasis on complex biological systems and reactive intermediates. Before she established the Computational Biochemistry group within the Faculty of Biology of at the University Duisburg-Essen, she was independent group leader at the Max Planck Institute for Coal Research (Mülheim). The Computational Biochemistry research group works on investigating the properties, reactive behaviour and applications of the most diverse systems, from reactive species to protein complexes. Implementation and application of hybrid methods, typically quantum mechanics/molecular mechanics approaches (QM/MM) and their combination with coarse-grained techniques (QM/MM/CG), are central to the research. For biomolecular applications, their studies facilitate predictions of binding sites in proteins and use this information for the development of improved ligands with tailored properties “in silico”. Meanwhile, mutations are also investigated as a tool for the regulation of biological processes. Foremost in this research is computer-assisted prediction of mutations for targeted modification of protein properties, e.g. of enzymatic activity. Another area of study is comprised by the role of intermolecular interactions between solvent and solute for the optimisation of enzymatic catalysis and for the control of protein aggregation and protein-ligand interactions. These are just some examples. Currently, the research group typically requires 25 million core hours per year, which are covered by magnitUDE, other clusters, and one application in Jülich. Prof. Sánchez-García is a member of significantly contributes of the Cluster of Excellence RESOLV (EXC 1069) on Solvation Science, of the Collaborative Research Centre 1279 and to the Collaborative Research Centre 1093 “Supramolecular Chemistry on Proteins”. CRC 1093 is a joint effort of researchers from the Faculties of Biology and Chemistry at the University of Duisburg Essen towards control of biomolecular function by supramolecular ligands.
